




Preview text:
VIETNAMESE NATIONAL UNIVERSITY – HCMC INTERNATIONAL UNIVERSITY
SCHOOL OF BIOTECHNOLOGY
MOLECULAR GENETICS
Laboratory Assignments (1, 2, 3) Group: 02 Group members: Nguyễn Thùy Dương BTBTIU18050 Nguyễn Nguyên Khang BTBTIU18098 Bùi Mỹ Yến Như BTBTIU18444 Nguyễn Việt Tiến BTBTIU18239 Course ID: BTBT18IU11 Instructor: PhD. Nguyen Minh Thanh Date of submission: May 25, 2021
LAB 1 – TEXT SEARCH FROM ONLINE DATABASES
Question 1: Explain the difference of gene structure from the searches of humsomi (Exercise 1) and
X59263 (Exercise 2).
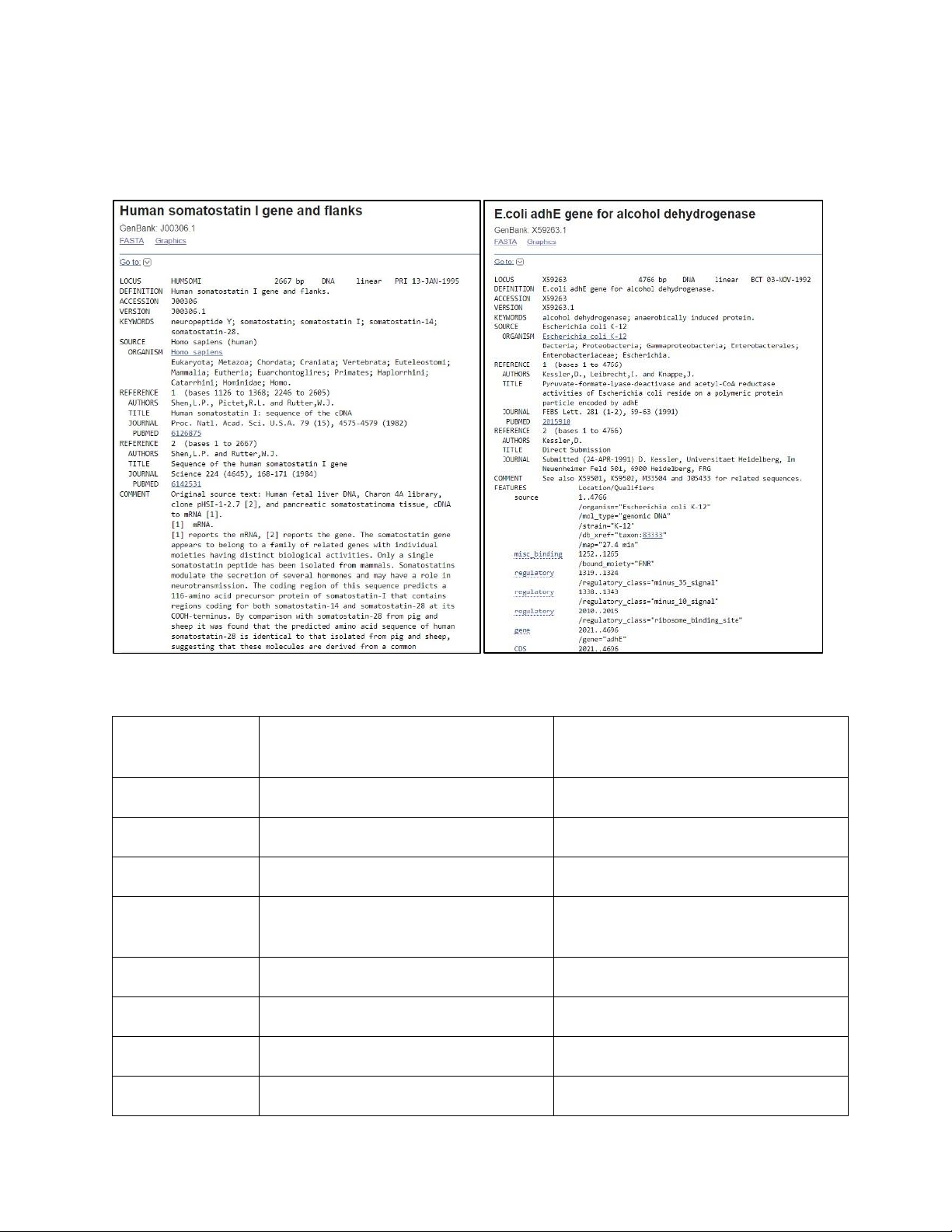
Image 1.1. Results gained from searching for “humsomi” and “X59263” on GenBank’s Nucleotide database.
Human somatostatin I gene and flanks E. coli adhE gene for alcohol dehydrogenase Locus name HUMSOMI X59263 Sequence length 2667 bp 4766 bp Molecular type DNA DNA GenBank PRI (primate sequences) BCT (bacterial sequences) Division Source Homo sapiens (human) Escherichia coli K-12 Exon 2 0 Intron 1 0 CDS 2 (1231..1368,2246..2458) 1 (2021..4696)
Question 2: Have a look through the results of Exercise 9, list species with MHC and without MHC
sequence identified. If there is species with MHC sequence, explain the reason. If there is species without
MHC sequence, try to explain the reason why it was not picked up.
- Species with MHC: Kangaroo (results shown under the scientific name Macropus eugenii),
gorilla, echidna (Tachyglossus aculeatus), sloth (Choloepus didactylus), tiger (Panthera tigris),
elk (Alces alces), dolphin (Delphinus capensis), cow (Bos taurus), elephant (results shown under
Elephas maximus and Loxodonta africana), tortoise (Gopherus polyphemus), domestic cat (Felis
catus), guinea pig (Cavia porcellus)
- Species without MHC: shark, panther
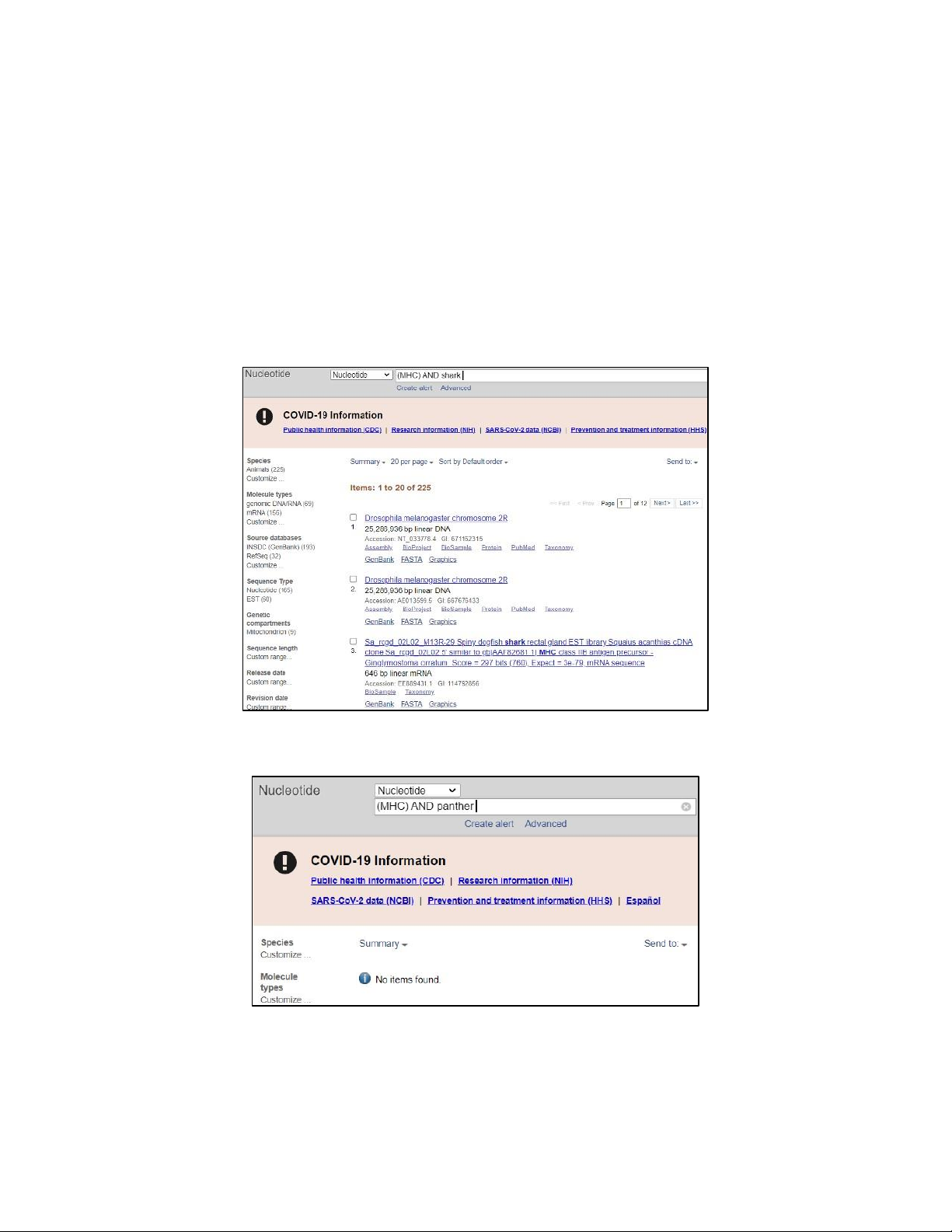
Image 1.2.a. Results found when searching for “MHC AND shark” are not closely related and obscured
on GenBank’s advance Nucleotide database.
Image 1.2.b. No result found when searching for “MHC AND panther” on GenBank’s advance Nucleotide database.
Since MHC (major histocompatibility complex) proteins are found in all higher vertebrates, it is
impractical to say that MHC sequence is not found in shark or panther. However, the searching for MHC
sequence of these species still shows unusual results. The reason to this might be the inaccurate searching
keyword – “panther” and “shark” – since there is no such Top organism listed in NCBI database as
“panther” or “shark”. If we want more accurate findings, we might want to use the specific scientific
name or family name of these species.
Question 3: Complete a search of NM003227 and answer the following points: -
Gene name: Homo sapiens transferrin receptor 2 (TFR2), transcript variant 1, mRNA - Gene length: 2886 bp -
Organism of origin: Homo sapiens (human) -
Number of introns: 0 (not reported). - Number of exons: 18
→ The number of introns is not reported from the search because this gene’s molecular type is mRNA.
Normally, introns are removed from pre-mRNA during RNA splicing while exons bonded together
covalently to create mature RNA. Therefore, introns are not expressed in the final mRNA product, thus
they are not reported while exons are presented in the searching of the sequence NM003227.
LAB 2 – SEQUENCE DATABASE COMPARISONS
Question 1: Using the results of the Exercise 2 explain whether a chromatin remodeling system is present in the insect.
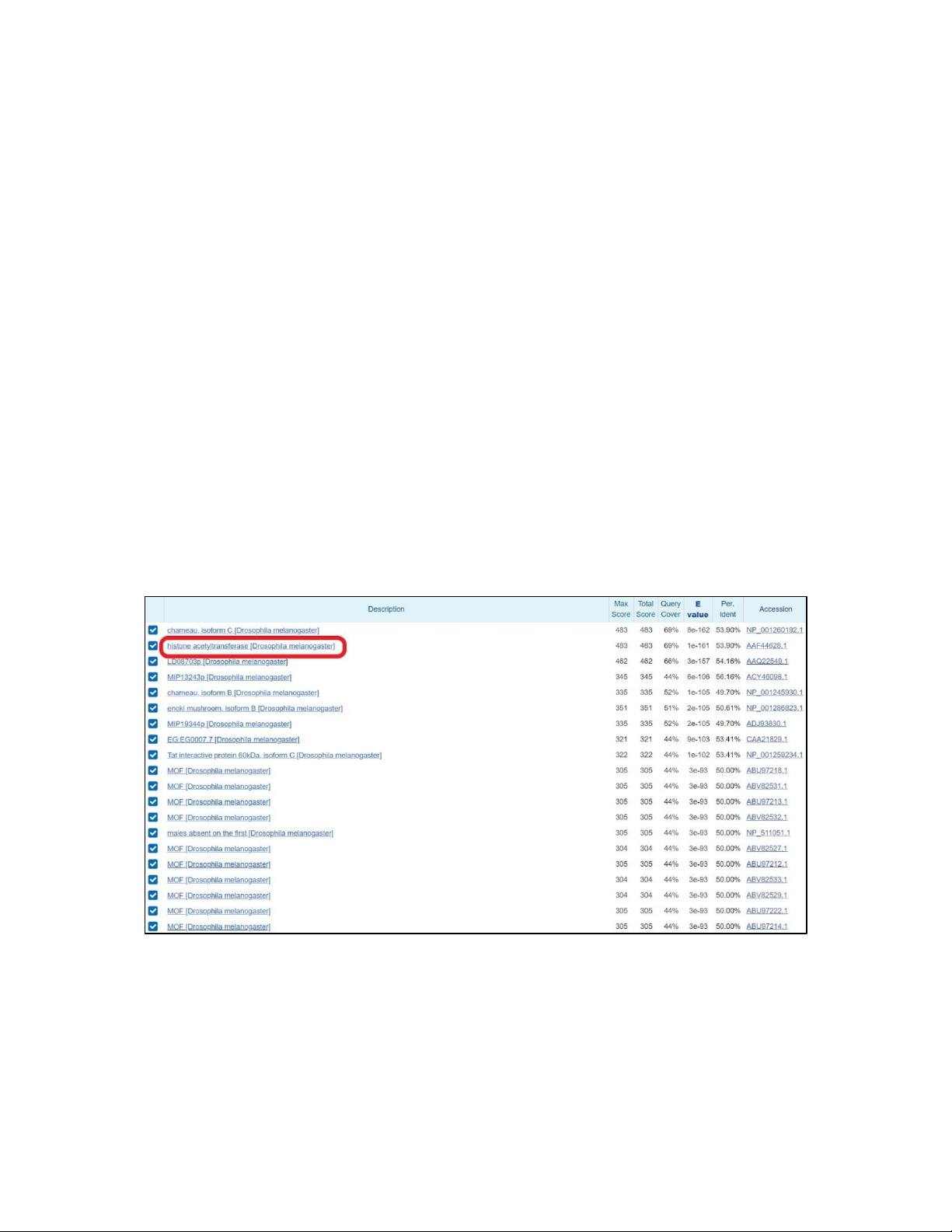
Image 2.1. Result gained from performing Blastp with the query sequence of histone acetyltransferase in Exercise 2.
A chromatin remodeling system is present in the insect since D. melanogaster was among the first
organisms used for genetic analysis, and today it is one of the most widely used and genetically
bestknown of all eukaryotic organisms. In eukaryotes, DNA is tightly wound into a complex called
chromatin. Thanks to the process of chromatin remodeling, this complex can be "opened" so that specific
genes are expressed. Chromatin remodeling is an important mechanism of regulating eukaryotic gene
expression, which makes tightly condensed DNA accessible to various regulatory factors, such as
transcription factors and components of DNA replication.
According to the results of BLAST, the identity levels range from 38-70%, which are mostly higher than
50%. So, the insects also have other types of proteins that have an identical sequence to histone
acetyltransferase in the human sequence is feasible and that identical can lead to similar functions.
Moreover, histone acetyltransferase, which also exists in Drosophila melanogaster, is classified as an
insect, as shown above. Therefore, there is highly possible that insects may possess a chromatin remodeling system.
Question 2: List the species do not have a homologous sequence with the query of Exercise 3. What may
you explain in term of evolution?
The species do not have a homologous sequence with the query of Exercise 3: • Nomascus leucogenys • Macaca fascicularis • Sapajus apella
Explanation: Evolution is defined as descent with modification from a common ancestor •
At the molecular level, the modification means changes in DNA and protein sequence, and
corresponding changes in protein function
• As mutations accumulate in sequences derived from an ancestral sequence, the derived sequences
diverge from one another over time, but sections of the sequences may still retain enough
similarity to allow identification of a common ancestry
Question 3: Set up Blast search for an unknown sequence in Blackboard. Identify potential gene for this
unknown sequence, its function and which organism carries this gene.
Image 2.3.a. Result gained from performing Blast search for given unknown sequence
This unknown sequence consists of the potential gene for Crustacean hyperglycemic hormone B
(CHH-B) known as crustacean hyperglycemic hormone gill form.
It possesses these following functions:
• Identified in gill, mandibular gland, and heart
• May act as a stress hormone
• May be involved in the control of MF secretion, molting, and reproduction
• Alternatively spliced in Macrobrachium rosenbergii (giant freshwater prawn).
Image 2.3.b. Detailed information of the gene code for Crustacean hyperglycemic hormone B (CHH-B),
the potential gene for the given unknown sequence.
LAB 3 – SEQUENCE ALIGNMENT
Question 1: Compare two protein sequences humins & ratins (GenBank EHB15713.1). What is their
identity, similarity, gap, and score? Used program: EMBOSS Water - Identity: 87/110 (79.1%) - Similarity: 94/110 (85.5%) - Gaps: 0/110 (0.0%) - Score: 457.0
Image 3.1. Alignment result of humins and ratins protein sequences using EMBOSS Water
Question 2: Align protein sequence humins with the common chimpanzee (Pan troglodytes) sequence
(GenBank NP_001008996). Report: the number of identical amino acids and the number of similar
physical/chemical amino acids. Used Program: EMBOSS Water -
Number of identical amino acids: 108 -
Number of similar physical/chemical amino acids: 109
Image 3.2. Alignment result between humins and common chimpanzee’s insulin protein sequences using EMBOSS Water
Question 3: Align actin gene of the white-leg shrimp (Litopenaeus vannamei) (GenBank AF300705) and
the giant freshwater prawn (Macrobrachium rosenbergii) (GenBank AY626840). Note the number of
transversion SNPs, the number of transition SNPs and their positions. Used Program: diffseq -
Number of transversion SNPs: 6 Location
Type of transversion SNPs
In white-leg shrimp’s gene
In gian freshwater prawn’s gene C A 258 225 C A 464 431 G C 494 461 T G 701 668 G C 935 902 A T 977 944 -
Number of transition SNPs: 2 Location
Type of transversion SNPs
In white-leg shrimp’s gene
In gian freshwater prawn’s gene C T 479 446 C T 827 794 - Total SNPs: 8
Image 3.3. Result gained from performing alignment of actin genes of the white-leg shrimp (Litopenaeus
vannamei) and the giant freshwater prawn (Macrobrachium rosenbergii) using diffseq
